Search options
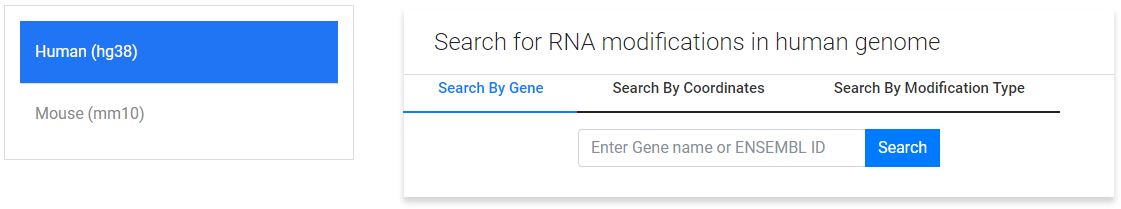
1. Gene based search:
Search format: "GENENAME - ENSEMBLID"
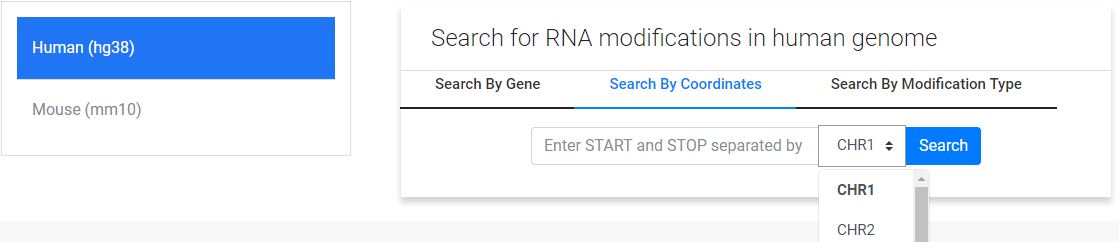
2. Coordinate based search:
Search format: "START|STOP"
3. Search by modification/study type:
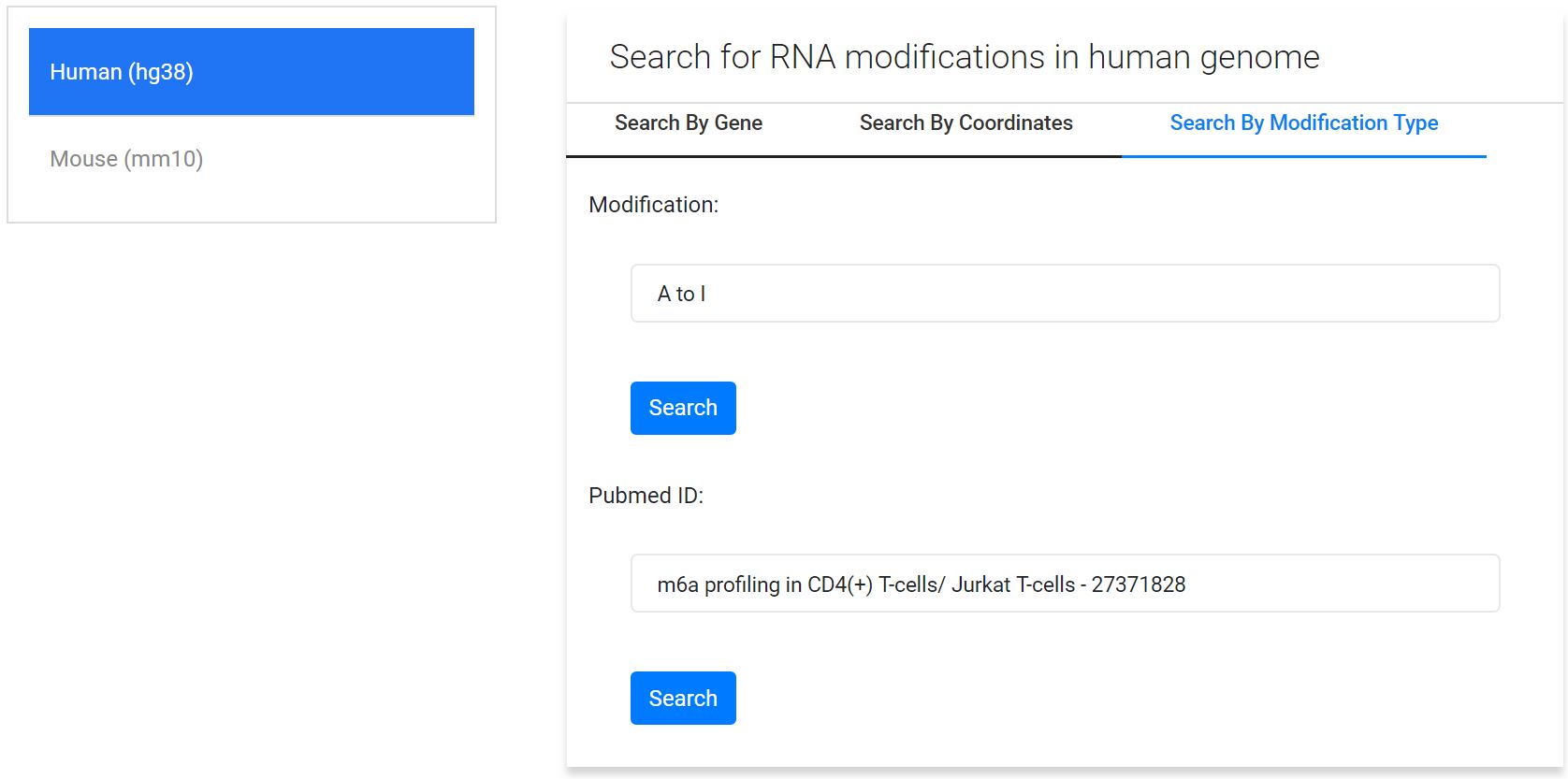
Result Page
1. Annotations:
For gene and coordinate based search, scrollable annotations providing more information on search query are displayed on top of the page

2. Data Tables:
The output is categorized by modification type into various tabs with number of results displayed next to modification name
3. Genome Browser:
3.a. Load custom BED/BAM/WIG/BIGWIG/BEDGRAPH files from URL
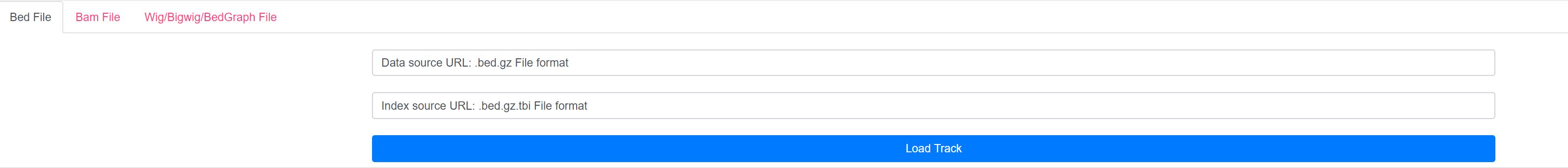
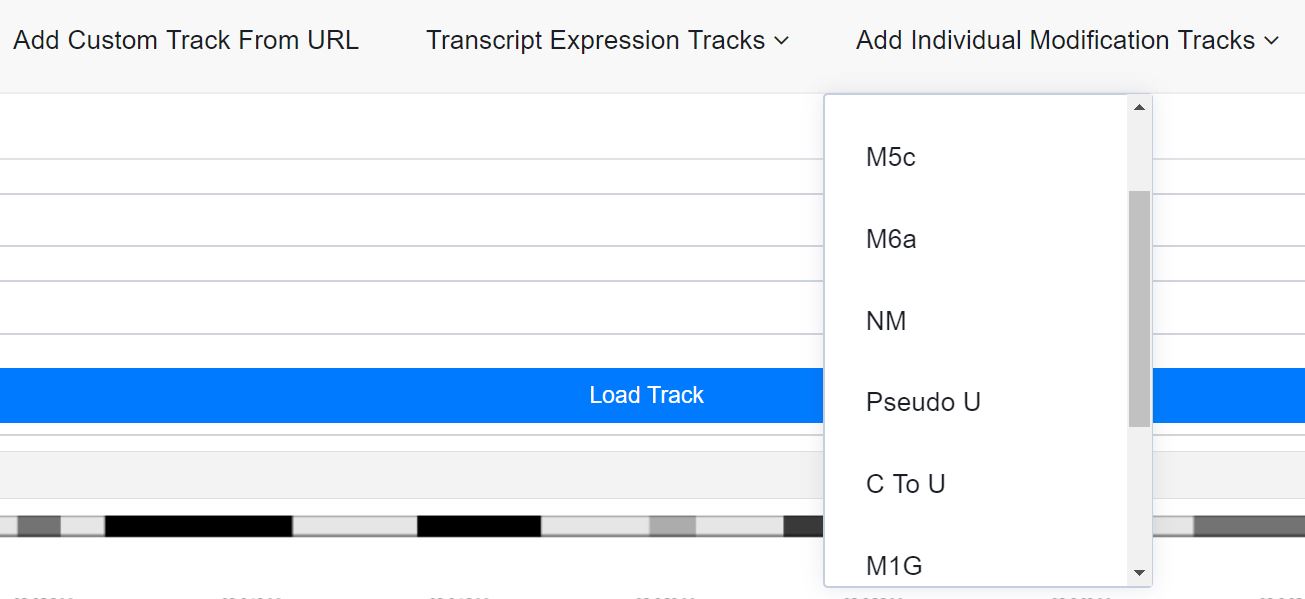
3.b. Load individual tracks for each expression level and for each modification
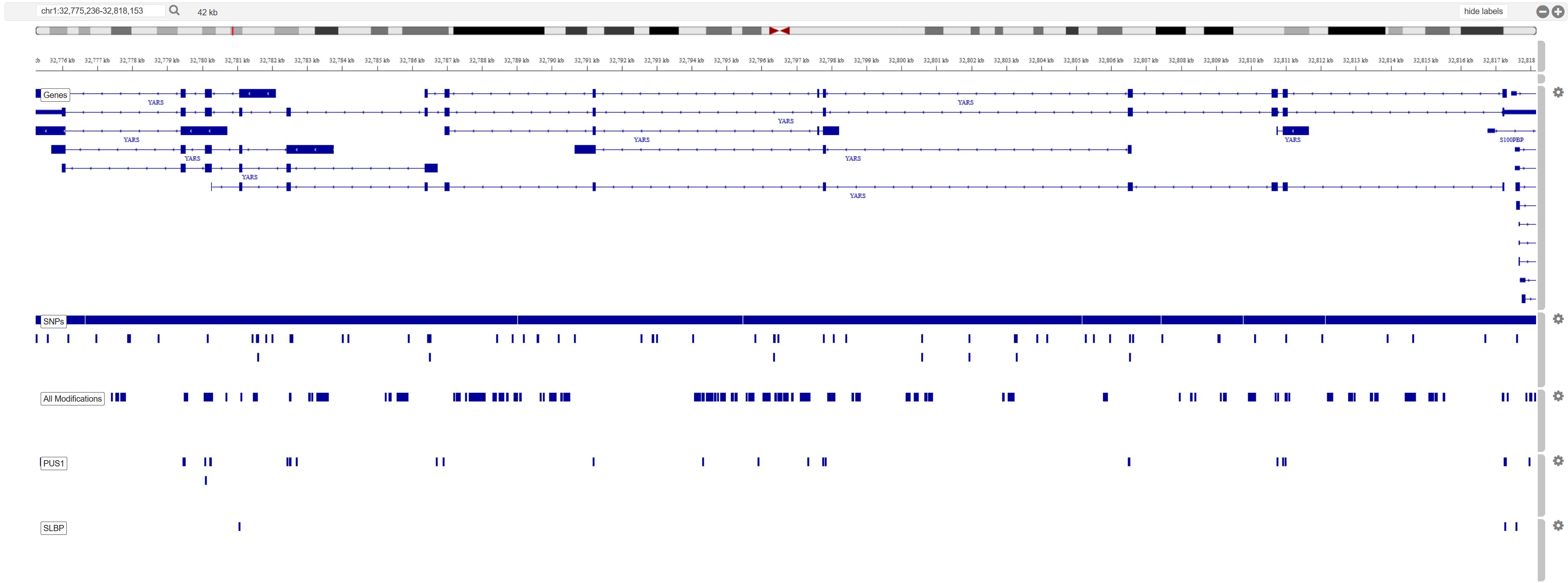
3.c. Browser view:
Genome browser by default loads base genome track, SNP track and a single track for all modifications
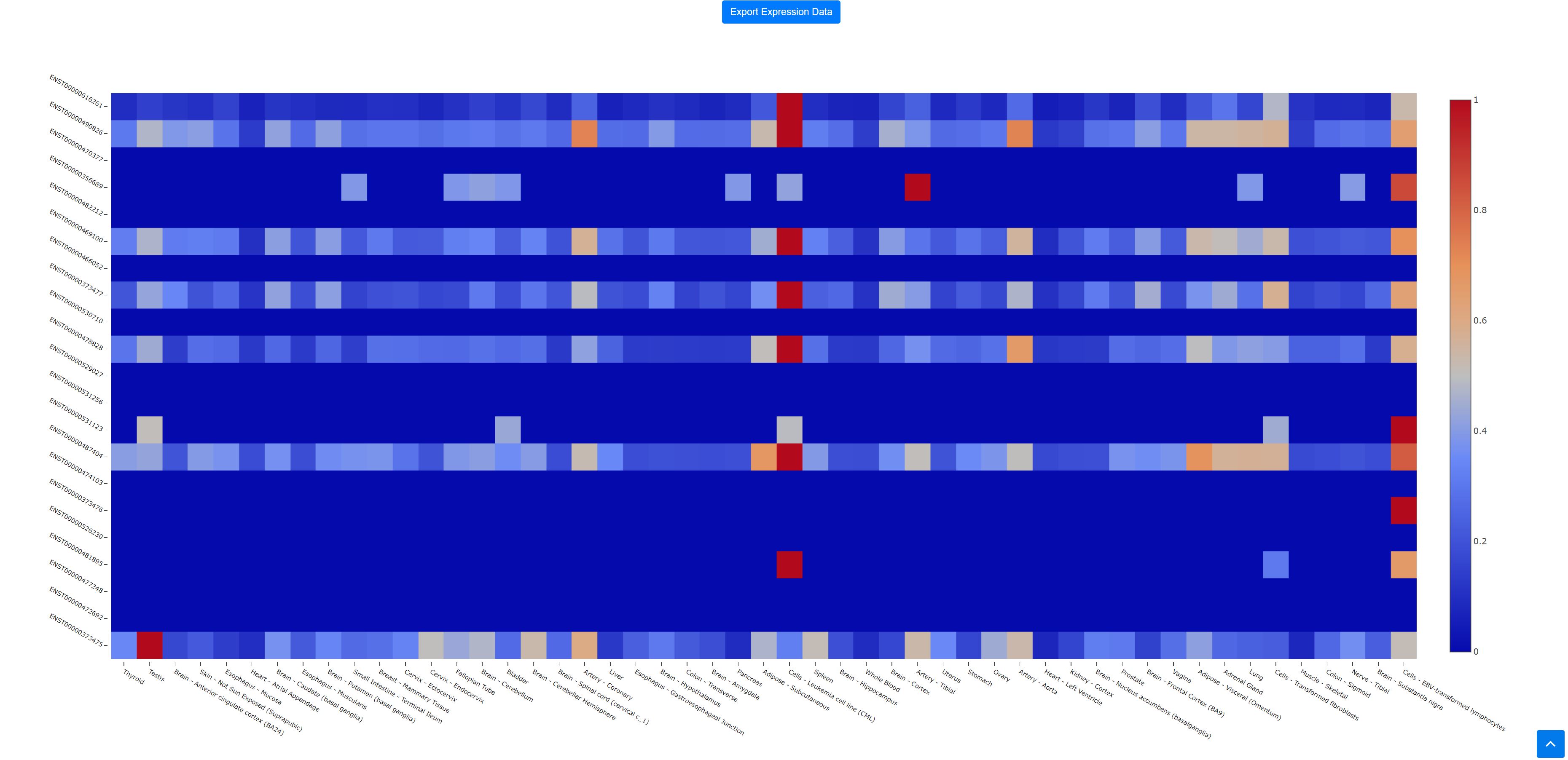
4. Transcript Expression Heatmap:
Heatmap can be accessed by opening the "View transcript expression tab" and the heatmap raw data can be downloaded. The data in the heatmap is normalized by row max however the mouse-over and download expression levels are actual values in RPKM.
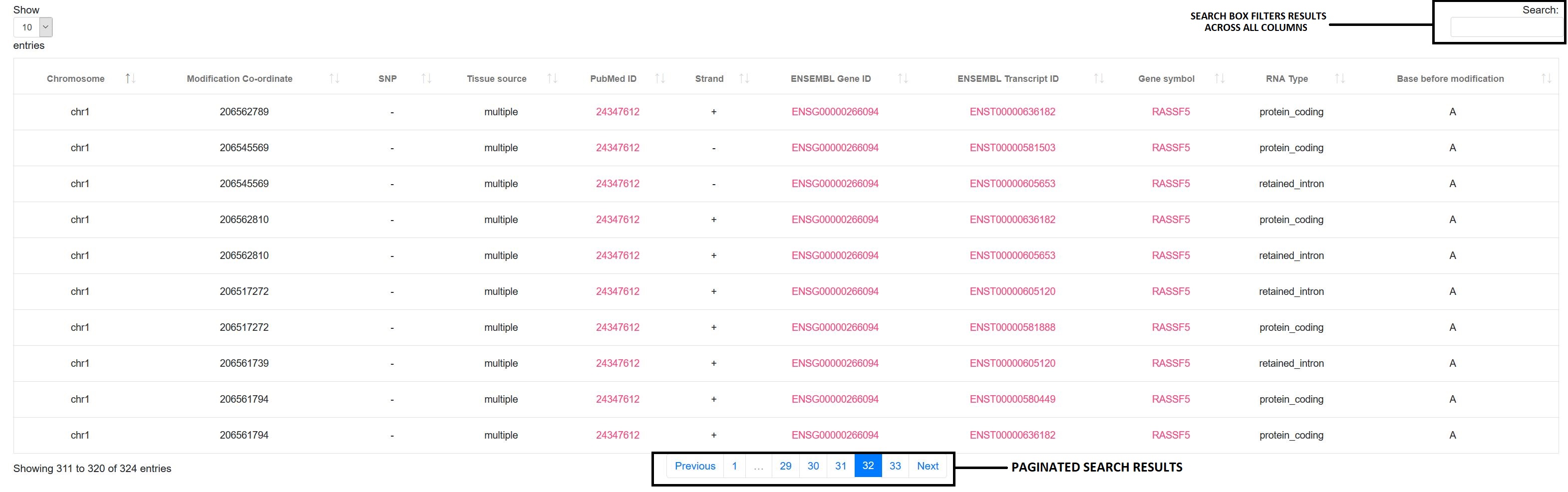
5. Modification based search:
If you have searched by modification, a simple table is displayed with 5 columns as depicted in the following image. This search method allows users to export data in various formats(Excel, PDF, print and csv)
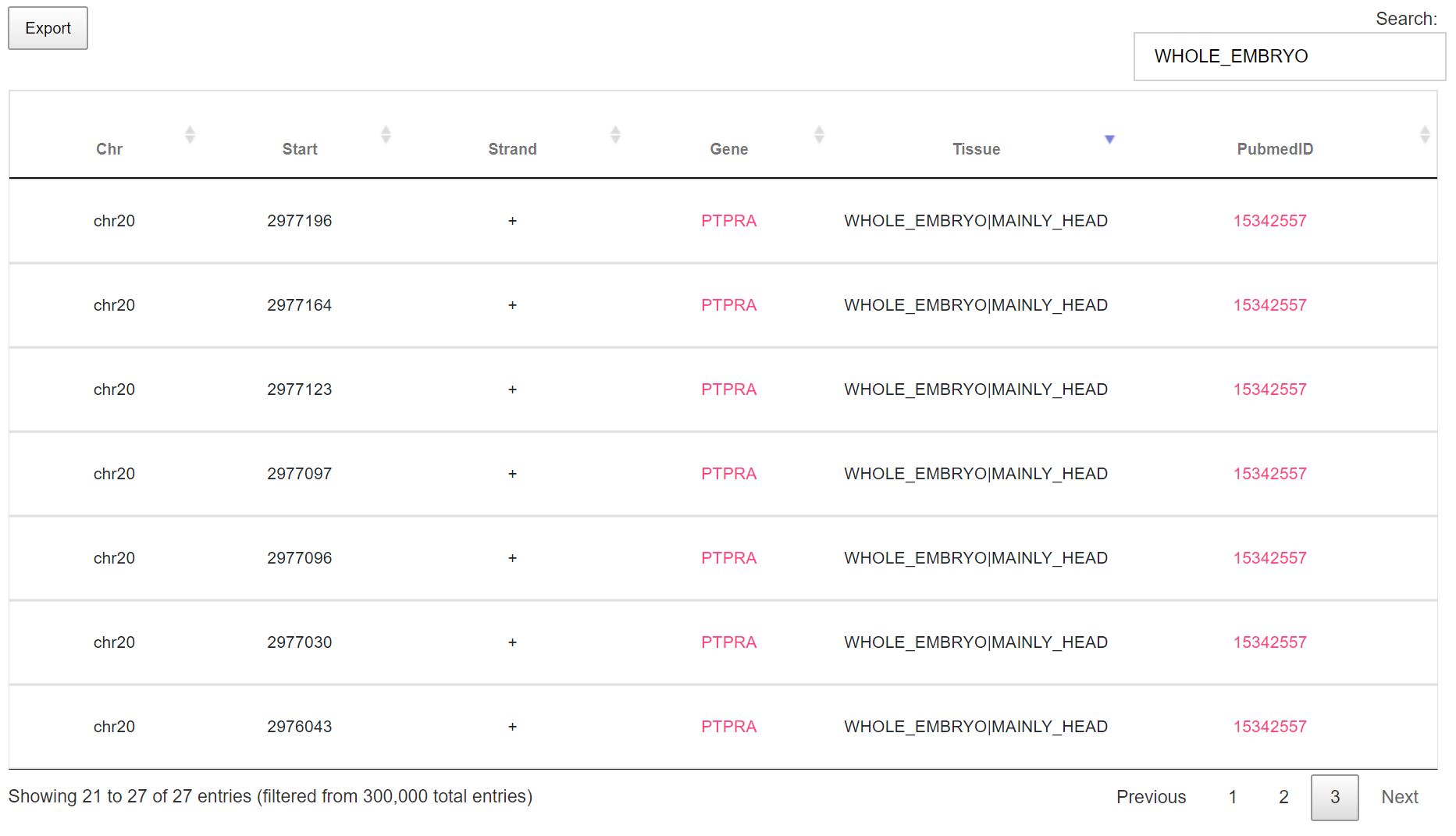
Key points:
-> On searching by modification type for A to I, m6A in humans and m6A in mice, the top 300,000 results are returned sorted by maximum hits for each gene.-> On searching by study type/Pubmed ID, the results retrieved will also include all other Pubmed IDs that support the modification type along with the queried Pubmed ID.
-> For coordinate based search, the maximum search range is restricted to 10,000 bases.